LDNFSGB: prediction of long non-coding rna and disease association
Background A large number of experimental studies show that the mutation and regulation of long non-coding RNAs (lncRNAs) are associated with various human diseases. Accurate prediction of lncRNA-disease associations can provide a new perspective for the diagnosis and treatment of diseases. The main function of many lncRNAs is still unclear and using traditional experiments to detect lncRNA-disease associations is time-consuming. Results In this paper, we develop a novel and effective method for the prediction of lncRNA-disease associations using network feature similarity and gradient boosting (LDNFSGB). In LDNFSGB, we first construct a comprehensive feature vector to effectively extract the global and local information of lncRNAs and diseases through considering the disease semantic similarity (DISSS), the lncRNA function similarity (LNCFS), the lncRNA Gaussian interaction profile kernel similarity (LNCGS), the disease Gaussian interaction profile kernel similarity (DISGS), and the lncRNA-disease interaction (LNCDIS). Particularly, two methods are used to calculate the DISSS (LNCFS) for considering the local and global information of disease semantics (lncRNA functions) respectively. An autoencoder is then used to reduce the dimensionality of the feature vector to obtain the optimal feature parameter from the original feature set. Furthermore, we employ the gradient boosting algorithm to obtain the lncRNA-disease association prediction. Conclusions In this study, hold-out, leave-one-out cross-validation, and ten-fold cross-validation methods are implemented on three publicly available datasets to evaluate the performance of LDNFSGB. Extensive experiments show that LDNFSGB dramatically outperforms other state-of-the-art methods. The case studies on six diseases, including cancers and non-cancers, further demonstrate the effectiveness of our method in real-world applications.

Pattern recognition analysis on long noncoding RNAs – a tool for

Prediction – lncRNA Blog

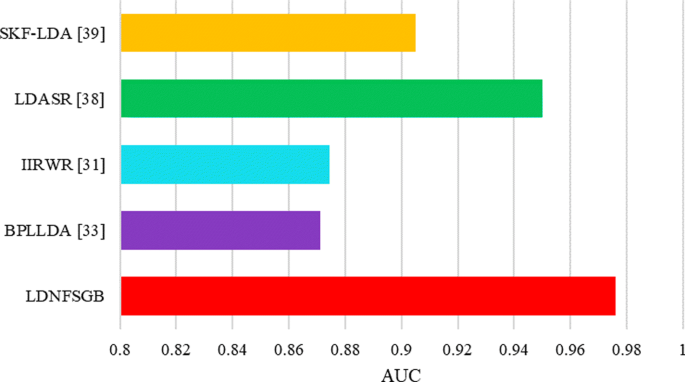
ROC curves of LDNFSGB for lncRNA-disease association prediction

RWSF-BLP: a novel lncRNA-disease association prediction model

The Pipeline of the LPI-HyADBS framework. (1) Initial feature
Life, Free Full-Text

Heterogeneous graph neural network for lncRNA-disease association

IJMS, Free Full-Text

Extra Trees Method for Predicting LncRNA-Disease Association Based

ROC curves of LDNFSGB for lncRNA-disease association prediction on
Frontiers Applications of Non-Coding RNAs in Patients With
PDF) LDA-VGHB: identifying potential lncRNA–disease associations

MAGCNSE: predicting lncRNA-disease associations using multi-view

ROC curves of LDNFSGB for lncRNA-disease association prediction